Our Research
We combine functional epigenomics with hands-on apiculture to dissect the molecular programs that transform a single genome in the honey bee into three distinct developmental outcomes.
The honey bee as a model for epigenetics
Honey bees (Apis mellifera) are haplodiploid organisms that live in complex societies comprising tens of thousands of individuals. Each colony contains two main diploid female castes: a single queen who is specialised for reproduction and thousands of sterile female worker bees. A third phenotypic outcome, which develops from unfertilized eggs, is a haploid male drone. Nutritional input during larval stages is the primary cue that sets differing post-embryonic trajectories and supports their maintenance in adulthood. We investigate how dietary signals are transduced into epigenetic and gene-regulatory programs to produce these divergent phenotypes.
Epigenetics of
caste determination
and male development
We characterise genome-wide cis-regulatory dynamics across the queen–worker caste polyphenism and the male (drone) developmental programme in Apis mellifera. Combining state-of-the-art epigenomic profiling, ATAC-seq, CUT&Tag/CUT&RUN, ChIP-seq, 3D genome conformation assays and WGBS, with high-resolution bulk and single-cell RNA-seq, and validating key nodes via targeted perturbations (CRISPR/RNAi knockdown, reporter assays), we reveal how regulatory programmes are converted into stable transcriptional states that specify queen, worker, or male fate.
Epigenetics and nutrition
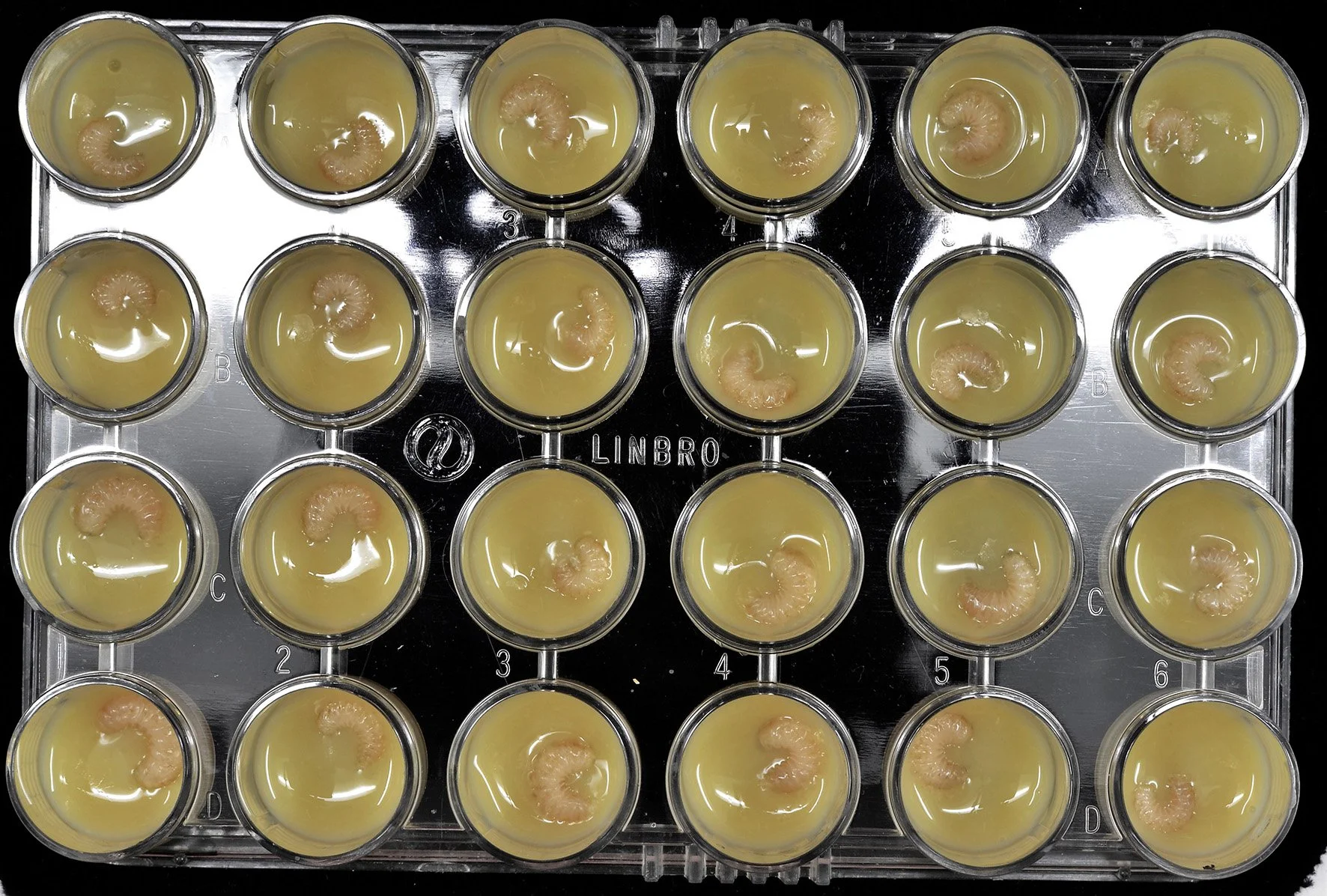
We manipulate larval diet in an in vitro growth pipeline (lab-reared, not in-hive) and quantify how nutritional inputs shape phenotypic outcomes, including queen–worker fate decisions and developmental rate. In parallel, we deploy heterologous systems to probe the functional components of royal jelly. Across both systems, we apply the same functional-genomics toolkit, ATAC-seq, CUT&Tag/CUT&RUN, ChIP-seq, 3D genome assays, WGBS, high-resolution RNA-seq, and targeted perturbations (CRISPRi/a, RNAi, reporter assays), augmented with metabolomics to profile nutrient and pathway states.